Mix infection analysis through machine learning
Mixed infections in genotypic drug-resistant Mycobacterium tuberculosis
Developed a custom Python pipeline using Gaussian Mixture Models to detect mixed M. tuberculosis infections from WGS data.
Analyzed over 5,000 public isolates to model expected allele frequency distributions and benchmark mixed infection detection thresholds.
Demonstrated that mixed infections are significantly associated with genotypic drug resistance, suggesting transmission complexity and diagnostic challenges.
Proposed integration of this method into existing TBProfiler workflows for improved strain-level resistance resolution.
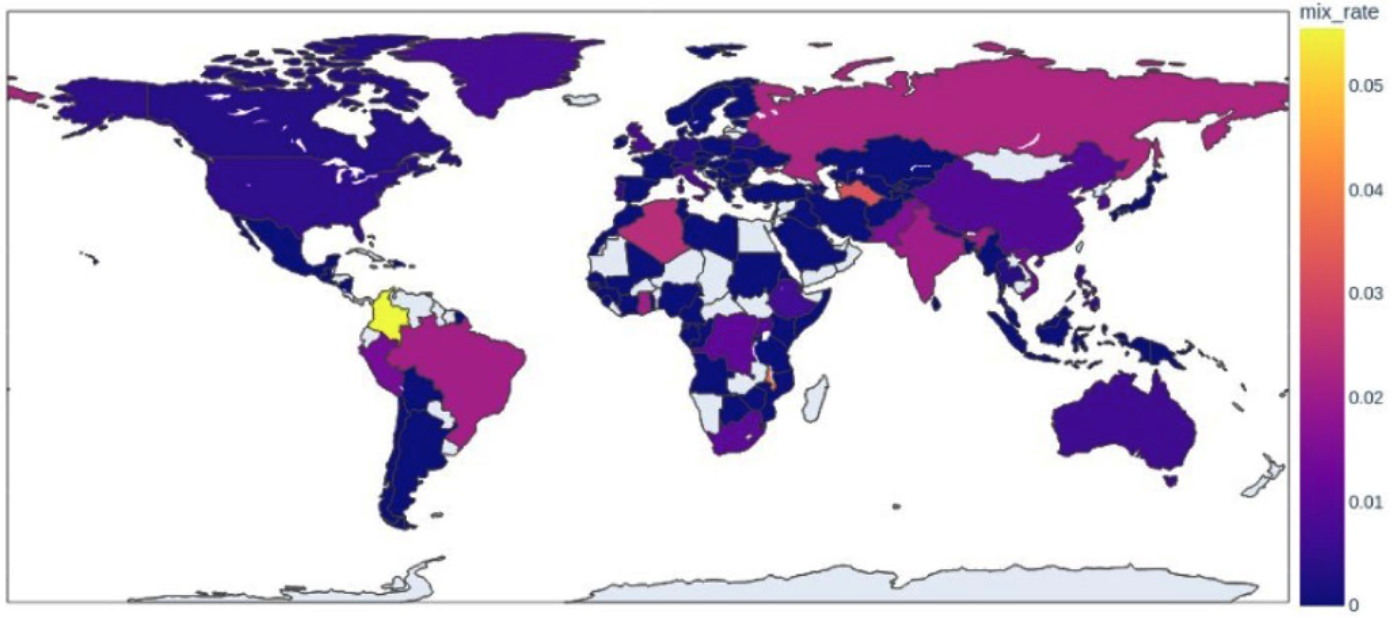
Mix infection over the world
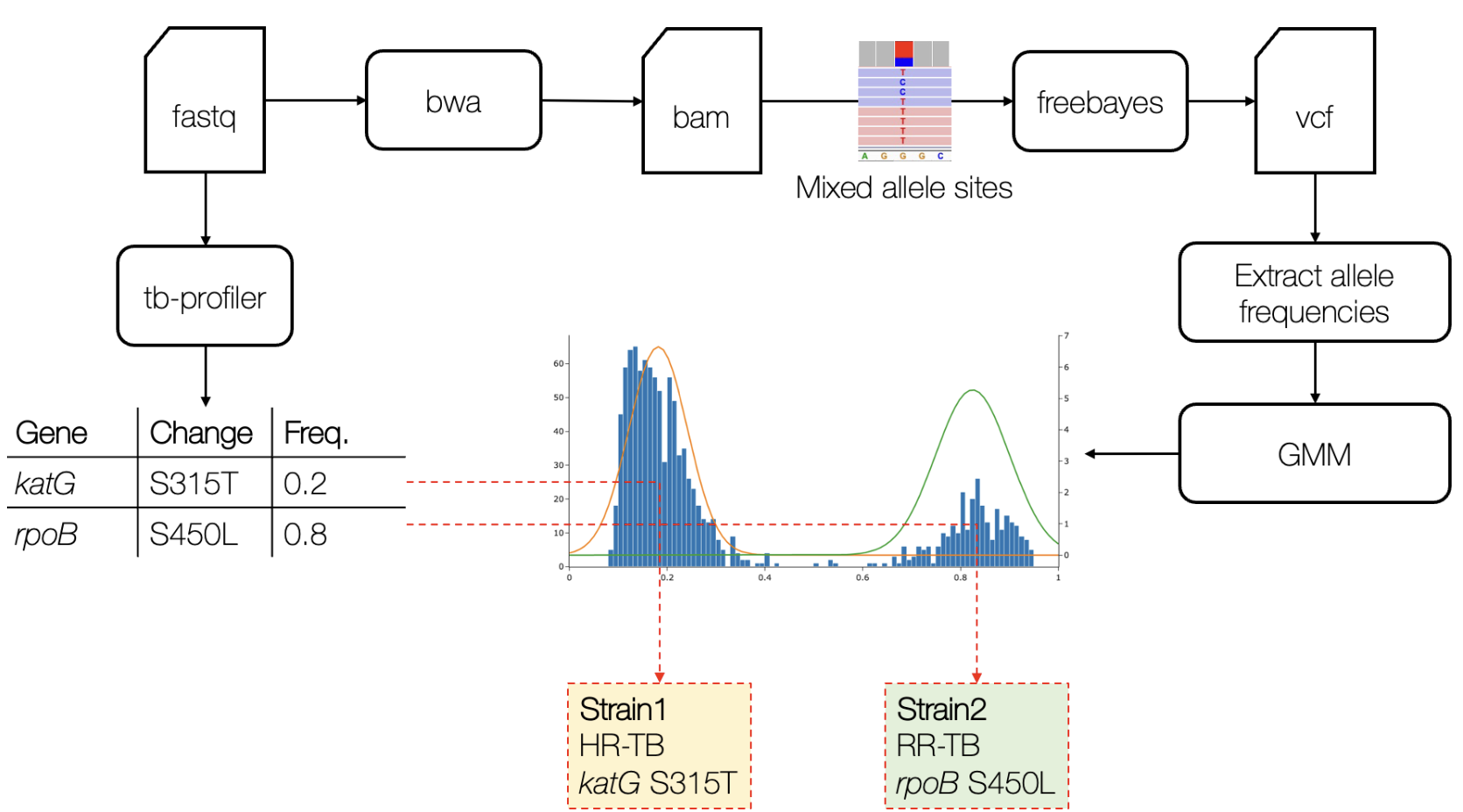
Mixed infection dissection pipeline